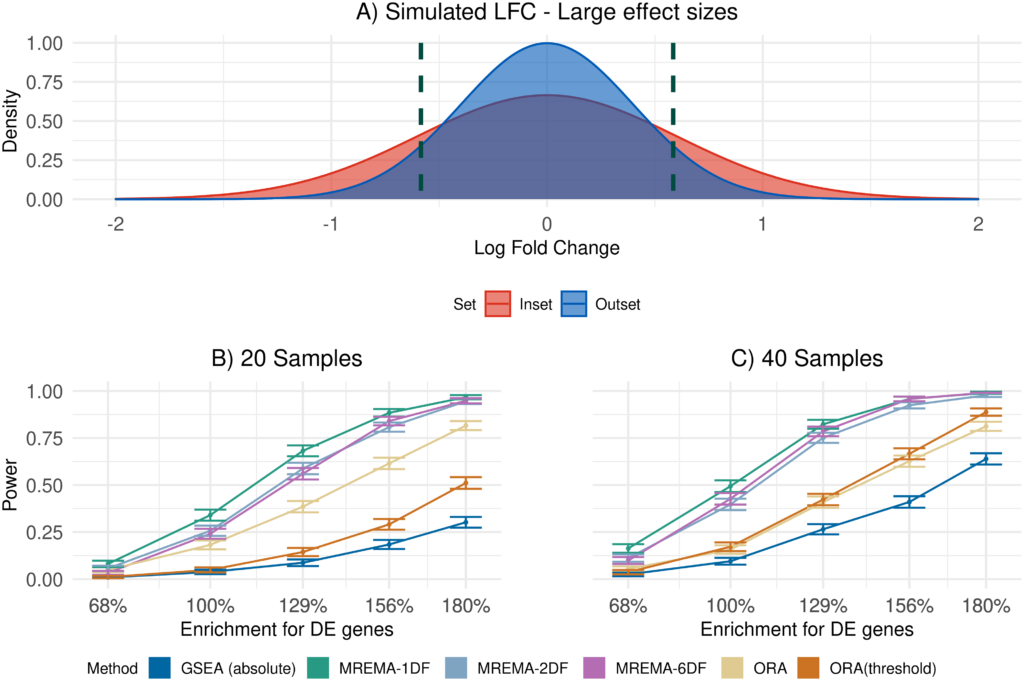
Well done to Donal O’Shea and his supervisor Cathal Seoighe on a recent publication “Random-effects meta-analysis of effect sizes as a unified framework for gene set analysis” in PLOS Computational Biology.

The role of gene set analysis is to identify groups of genes that are perturbed in a genomics experiment. There are many tools available for this task and they do not all test for the same types of changes. Here the authors proposes a new way to carry out gene set analysis that involves first working out the distribution of the group effect in the gene set and then comparing this distribution to the equivalent distribution in other genes. Tests performed by existing tools for gene set analysis can be related to different comparisons in these distributions of group effects. A unified framework for gene set analysis provides for more explicit null hypotheses against which to test sets of genes for different types of responses to the experimental conditions. These results are more interpretable, because the group effect distributions can be compared visually, providing an indication of how the experimental effect differs between the gene sets